[Day 191] Starting the book - Effective Data Science Infrastructure
Hello :)
Today is Day 191!
A quick summary of today:- today I started a book from Manning - Effective Data Science Infrastructure
Below is a summary of the topics covered up to and including chapter 3.
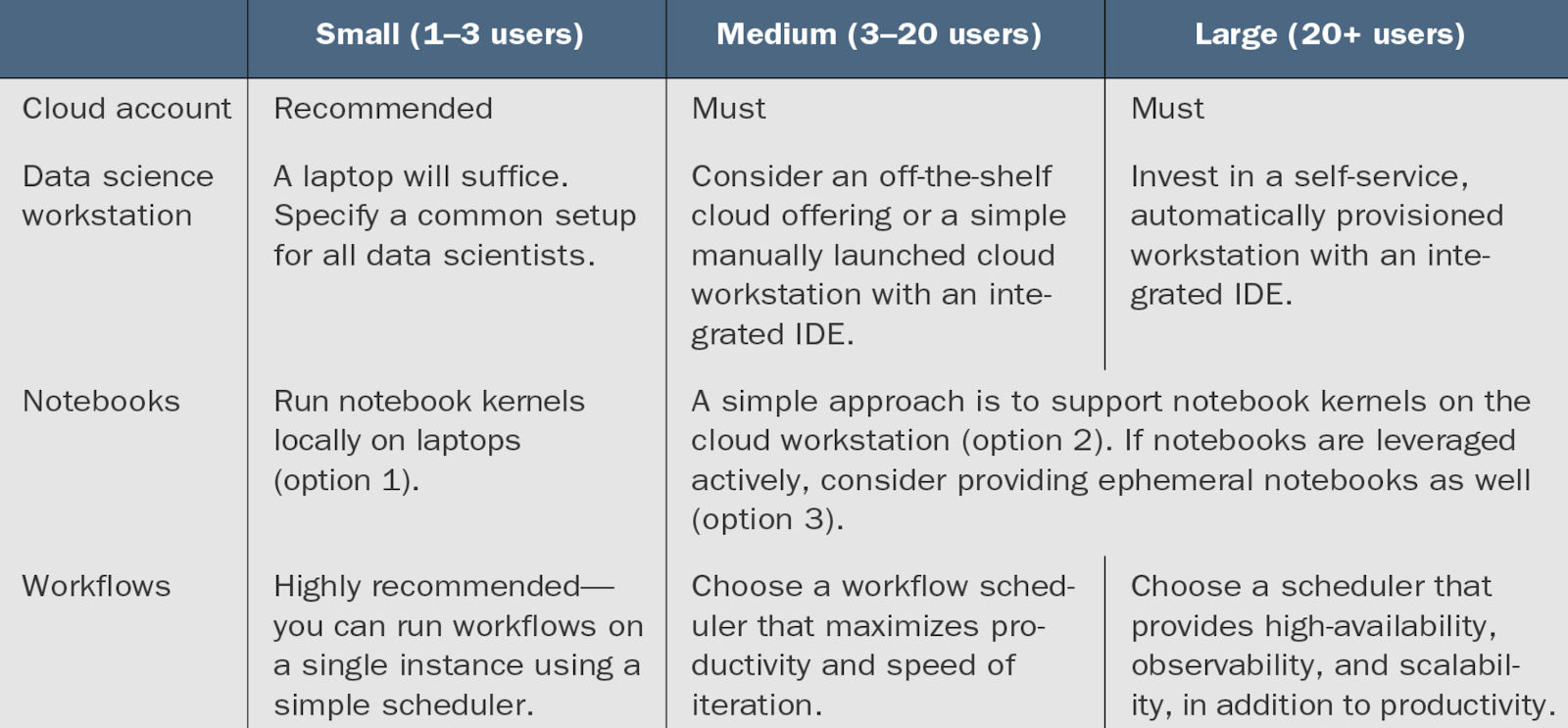
To enhance data scientists' productivity, prioritize a well-structured development environment over solely focusing on scalable production infrastructure. Many companies overlook this, managing code development, debugging, and testing haphazardly. Effective development environments, integral to infrastructure, enhance productivity by focusing on ergonomics, defined as optimizing efficiency in the working environment.
Key activities to optimize include:
1. Prototyping - Translating expertise into functional code and models.
2. Interaction with Production Deployments - Connecting code and models to systems to generate business value.
The prototyping loop (similar to the REPL loop in software engineering) involves developing, evaluating, and analyzing code iteratively. To boost productivity, streamline each step and transition in this loop.
This prototyping loop integrates with the higher-order loop of production deployment, forming an infinite cycle of improvement and debugging, akin to continuous delivery (CD) in software engineering. However, data science differs from traditional software in:
1. Correctness - Validated post-deployment through experiments, not pre-deployment tests.
2. Stability - Affected by constantly changing data, unlike stable software environments.
3. Variety - Requires iterative testing with new ideas and data, unlike static software functions.
4. Culture - Data scientists are not typically trained in the deep culture of DevOps and infrastructure engineering, necessitating a human-centric approach to development environments.
Running notebooks (options)
Example workstation for a data scientist
Workflows
a workflow is a directed graph, that is a set of nodes or steps connected by directional arrows
executing workflows is the job of an orchestratorWhen evaluating orchestrators, we can think of the architecture, which includes the code structure and system interface; the job scheduler, which involves workflow triggering, execution, monitoring, and failure handling; and compute resources, which consider the system's ability to handle varying resource requirements and parallel execution capabilities.
When choosing a workflow framework for data science, prioritize the productivity of data scientists by selecting an architecture tailored to data science use cases and considering the full data science stack. Operational concerns like robustness, scalability, and high availability are critical long-term factors, so opt for systems with proven reliability. Ensure the framework integrates seamlessly with your compute resources to enhance productivity.This book uses metaflow and I hope they provide clear instructions on setting it up.
What dev env is best for your team size?
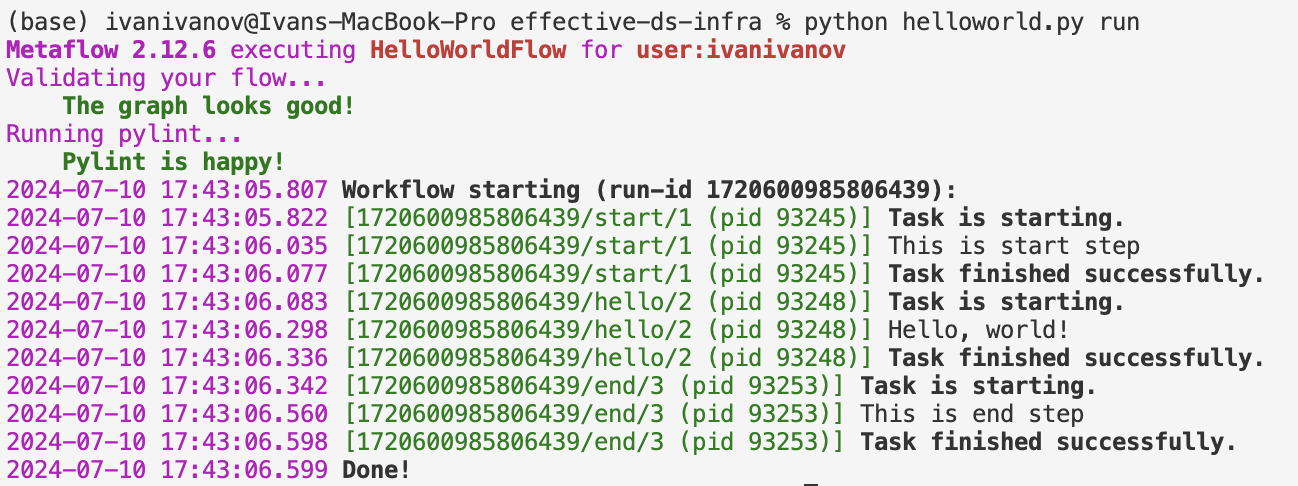
First, install with pip install metaflow
Then create a hello world flow:
Its output when run:
when we run the file with `show` added in the terminal, we see a clearer flow
this is a textual representation of the DAG
when we execute the flow, we see:
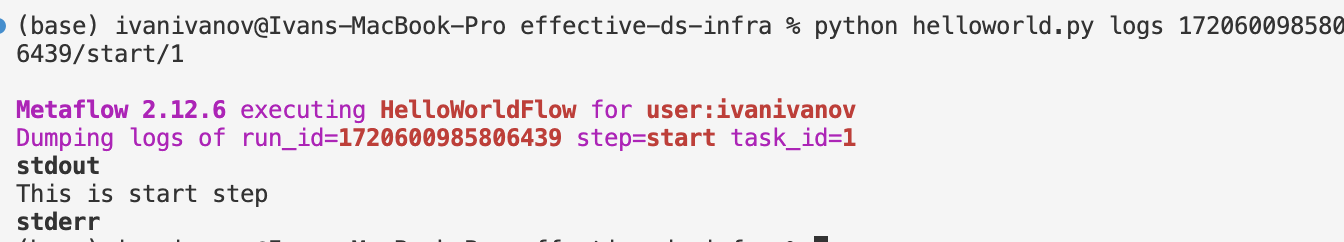
Using the run id, we can check the flow that was executed:
Next, metaflow artifacts
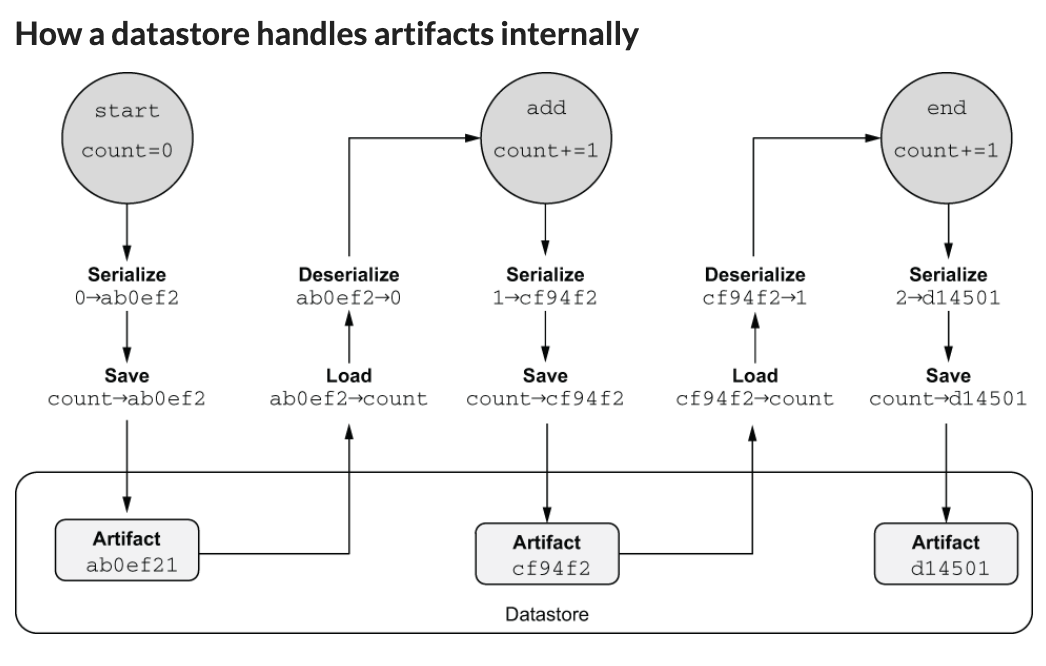
Metaflow artifacts (seems they are like artifacts in MLflow) are data objects automatically persisted by Metaflow when assigned to self in step code. They can include any Python objects serializable with pickle. Artifacts are stored in a datastore for reproducibility, maintaining immutability for accurate experiment tracking across workflow steps.
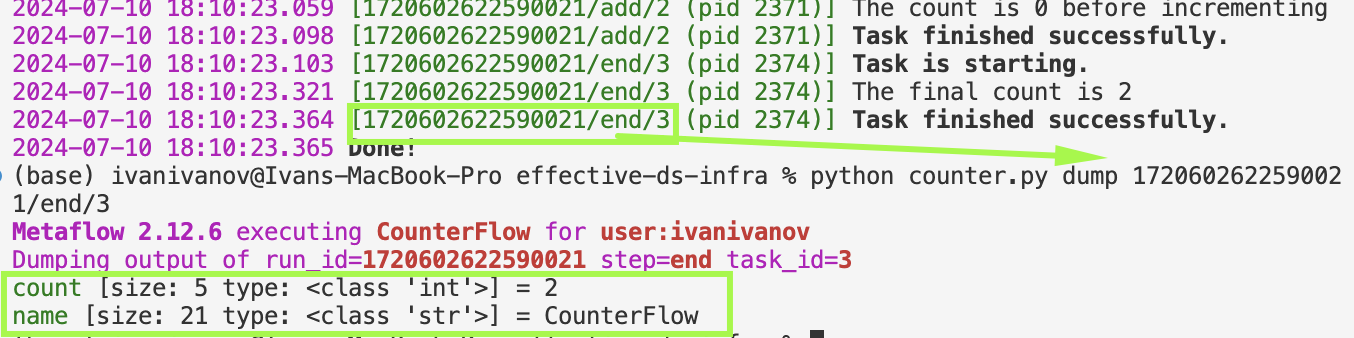
To illustrate artifacts, we created this simple CounterFlow
RULE OF THUMB
Use instance variables, such as self, to store any data and objects that may have value outside the step. Use local variables only for intermediary, temporary data. When in doubt, use instance variables because they make debugging easier.
Flows may fail. Because of that metaflow saves each state of the flow prior to the failure - and by examining this we can better understand what happened.
When we run the flow, we get the run id and step, and we can examine the state at that point
how the metaflow datastore works:When designing my flows in Prefect, for my last project I had thought a lot about what to put in a step - do I have multiple things, do I make it simpler? And usually I sided to put less functionality in 1 single step - so that if a step fails I know exactly where it fails. Whereas if a lot of operations were in a single step it might require more debugging. And the book provides a similar rule of thumb:
Structure your workflow in logical steps that are easily explainable and understandable. When in doubt, err on the side of small steps. They tend to be more easily understandable and debuggable than large steps.
Next - parameters
Parameters are a flow level construct and are available for all the steps in the flow
We can create params using the Parameter class
The actual flow is printing info about the params, and when we do just python parameters.py run
we get:
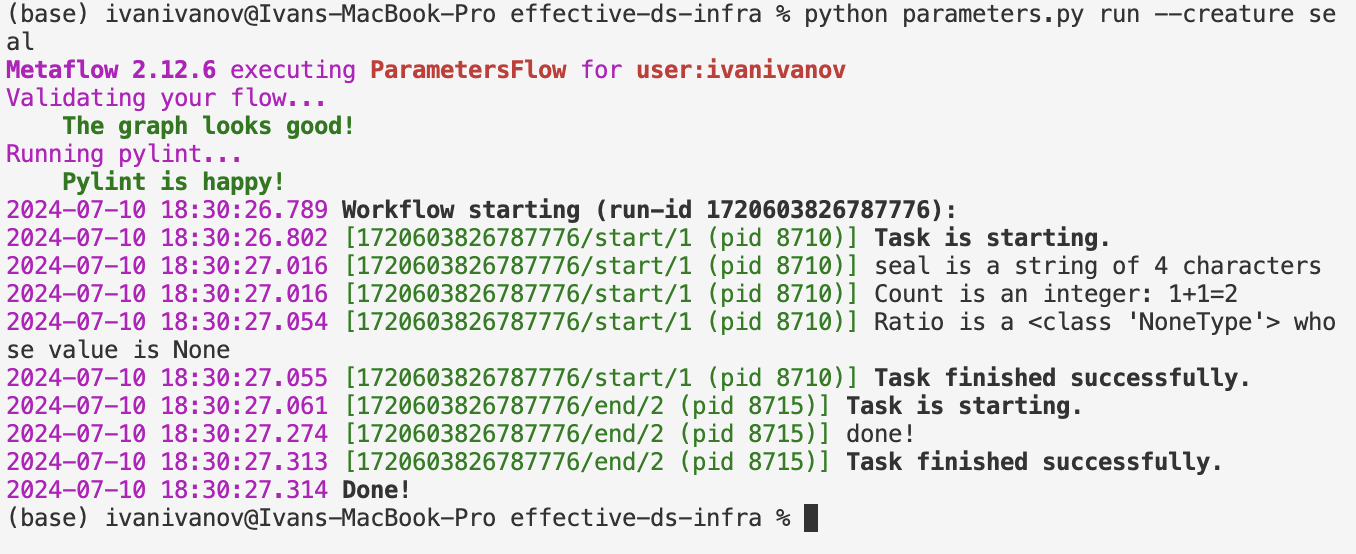
so if we pass creature like: `python parameters.py run --creature seal`
we get:
And the rest of the params can be passed the same way but they are not requiredParams are constant immutable values and if they need to be changed in the code, we need to create a copy and assign it to another artifact.
we can also set env vars to be used in runs. Like: `export METAFLOW_RUN_CREATURE=dinosaur` and this value will automatically be used instead and we don’t need to pass —creature when we run the flow
Even if a param is set in the env vars, if we pass —creature new_creature then this will be used and has priority
We can also add more complex params (not just int/float/str) using json:
json file paths can also be passed using —mapping like: `python json_parameter.py run --mapping "$(cat myconfig.json)"`
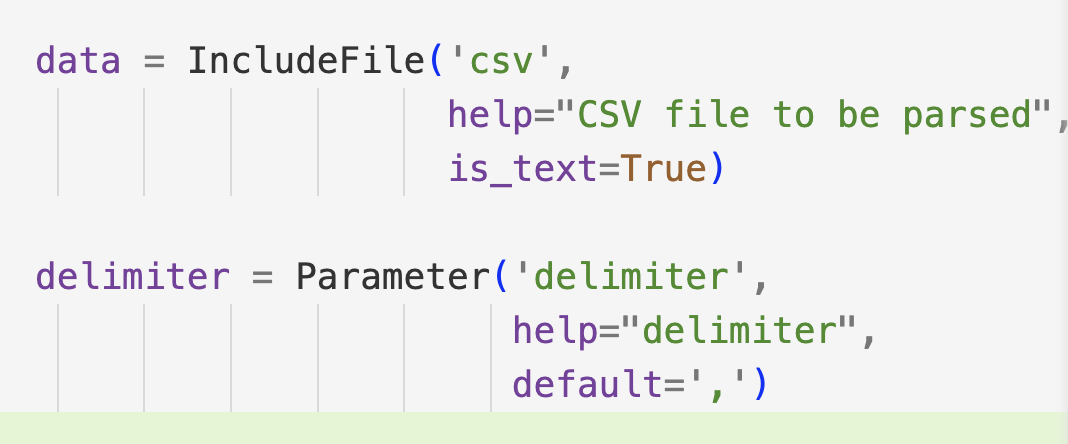
To include files as params, we can use the IncludeFile class from metaflow, and to add something like csv files as params we can use help="delimiter", default=',' when defining a param like:
In Metaflow, IncludeFile and delimiters serve distinct purposes:
IncludeFile: Reads a file and stores it as an immutable artifact with the run. This means the original data is captured and versioned along with the run, ensuring reproducibility even if the source file changes or is removed.
Delimiter: Specifies how fields in a CSV file are separated (e.g., comma-separated values). This ensures proper parsing of the CSV file's content.
So, while IncludeFile preserves data integrity by snapshotting it with the run, delimiters ensure correct parsing of CSV data into fields.
Running dynamic branches and data parallelism - can be done using the `foreach` keyword where a step is run for multiple cases in parallel
foreach can be used to execute and run in parallel even tens of thousands of tasks - it is a powerful keyword and a key element that contributes to scalability that metaflow provides.
To protect users from over-running tasks and making their machine heat up, metaflow has a preset concurrency limit
There are also two variables that help: max_num_splits controls data partitioning for parallel processing, while max_workers limits the number of concurrent executions to manage system resources effectively during parallel execution in Metaflow workflows. Adjusting these parameters can optimize performance based on specific computational needs and available resources.
Next, staring a simple classifier prediction model flow:
After running the flow, we can inspect the logs for the start step and see the artifacts:
(we can see at least some of it, and ensure it is loaded)
Since I ran the flow once, these artifacts are persisted and next time instead of loading them again from the API, I can load them from the metaflow datastore.
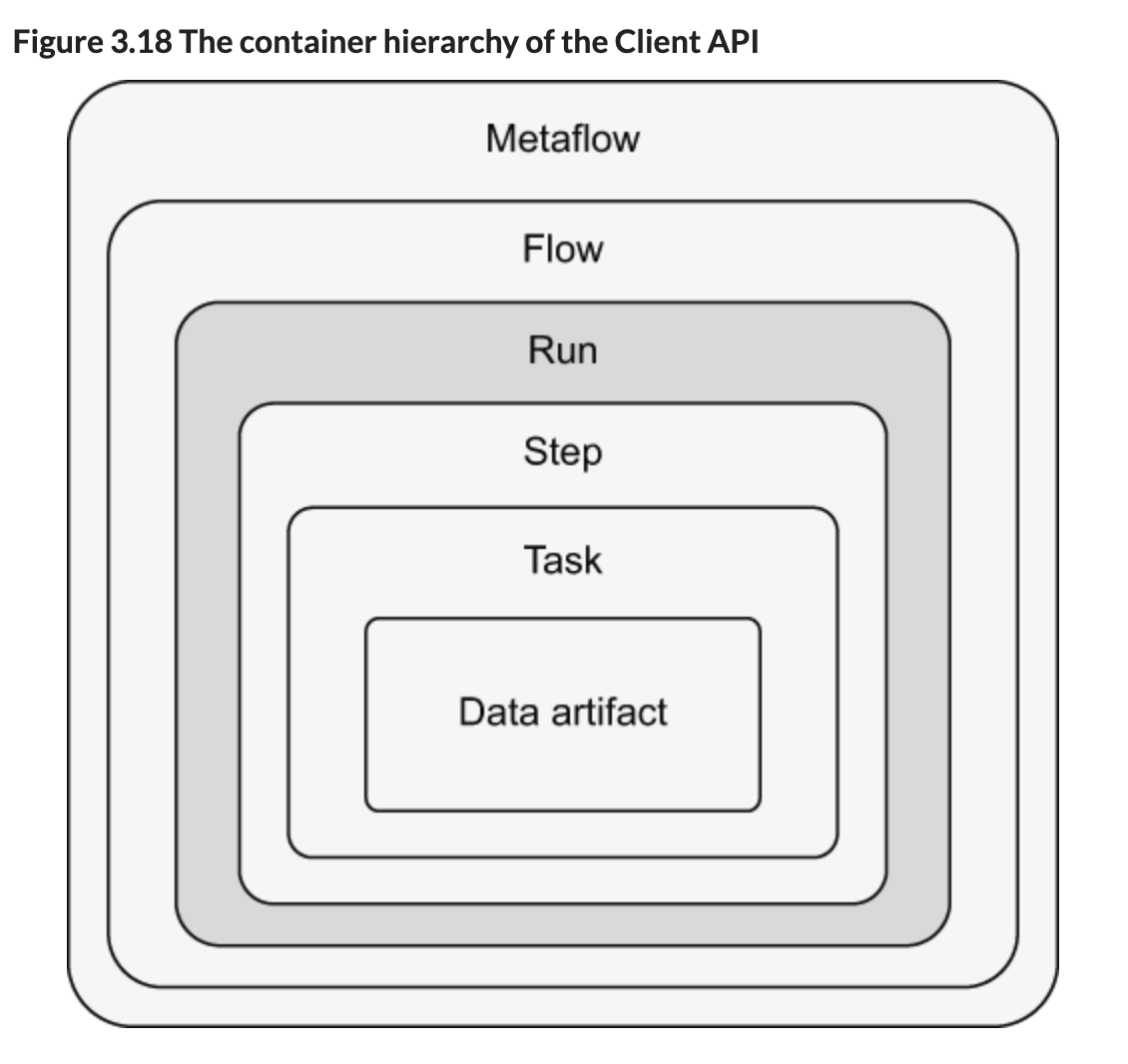
I am writing this as I go, and now I see a nice definition of flow, step, task, etc:
Metaflow—Contains all flows. You can use it to discover flows created by you and your colleagues.
Flow—Contains all runs that have been executed with a FlowSpec class.
Run—Contains all steps of a flow whose execution was started during this run. Run is the core concept of the hierarchy, because all other objects are produced through runs.
Step—Contains all tasks that were started by this step. Only foreach steps contain more than one task.
Task—Contains all data artifacts produced by this task.
Data artifact—Contains a piece of data produced by task.
To load a file there are 3 ways:
Instantiating Objects: You can create any object using a path that uniquely identifies it in the structure. For example, Run("ClassifierTrainFlow/1611541088765447") accesses the data of a specific run.
Accessing Child Objects: Using bracket notation (['start']), you can retrieve specific child objects. For instance, Run("ClassifierTrainFlow/1611541088765447")['start'] gives you the start step of that run.
Iterating over Containers: You can iterate over any container to access its children. For example, list(Run("ClassifierTrainFlow/1611541088765447")) returns a list of all step objects associated with that run.
And we can access the client API anywhere where python is supported - we can load files in another place like a jupyter notebook too.
When running a flow, if the flow fails for some reason, we can inspect - fix the big, and continue that specific run from the last successful step using python file.py resumeresume also accepts a specific run id and a step from which to resume
from chapter 3, the final full DAG looks like:
1. We obtained input data and split it into a train and test sets, which were stored as artifacts.
2. We trained two alternative models as parallel branches . . .
3. . . and chose the best performing one based on the accuracy with test data.
4. The chosen model was stored as an artifact, which we can inspect together with other artifacts in a notebook using the Client API.
5. A separate prediction flow can be called as often as needed to classify new vectors using the trained model.
Overall - very nice. After failing to setup metaflow (its UI to be specific) some days ago, following this chapter so far I had no issues and I can easily learn about metaflow. I hope the next chapters are just as good.
All code from today is on my repo.
That is all for today!
See you tomorrow :)