[Day 79] Attempting to make a Local Retrieval Augmented Generation (RAG) from Scratch
Hello :)
Today is Day 79!
A quick summary of today:- Started this tutorial for building a local RAG from scratch and dealt with issues due to me being on a mac
The tutorial is focused on cuda, but instead of colab, I wanted to try to do this locally:
1. to see the limits of my m2 pro
2. to deal with issues not shown in the video
I am almost at the end - 4:49:34 / 5:40:58, but it is 2am and I am falling asleep on the keyboard even writing this post now.
The tutorial is pretty amazing, it follows the below graph
The pdf used for the tutorial is a 1200 page book about nutrition, and the goal is to create a model with which we can talk and ask questions about nutrition. My thoughts of using this are afterwards I would use research papers, and test it with them.The steps to success are:
- Open a PDF doc (or collection of PDFs)
- Format the text, ready for an embedding model
- Embed the chunks of text and turn into embeddings
- Build a retrieval system that uses vector search to find relevant chunks based on a query
- Create a prompt that incorporates the retrieved pieces of text
- Generate an answer to a query base on the passages of the textbook with an LLM
Steps 1-3 resulted in
As for the embedding model, all-mpnet-base-v2 from huggingface is used.Another great thing about the tutorial is that the teacher just gives resources left and right. https://huggingface.co/spaces/mteb/leaderboard < this for example is a leaderboard of embedding models, and I can use anyone of them depending on my goal and resources.
For the embeddings, two methods were shown, raw, and in batches which is the fastest
Because I am running everything locally, it was slower for me than the teacher with an expensive GPU haha, and after I ran the 1st one for a few secs I saw that compared to the tutorial it is incredubly slow, so I just stopped it and ran the batch one. However, later I noticed that 95% of my embeddings in my df were nans. This was because the result from the 1st code was used, and not the embeddings from the batch run. I wanted to note it to the teacher, and I raised an issue on github so that other students can see it in case they run in a similar issue, which I regret - I could have made it a discussion or just left a youtube comment (and deleting is not allowed). It is what it is ~ I fixed that for my case and moved on.The next issue I ran into is, because the df is converted and saved to a csv, and then loaded to be used. In that conversion the embedding column is turn to strings. In the video it is covered how to do it, with:
text_chunks_and_embeddings_df['embedding'] = text_chunks_and_embeddings_df['embedding'].apply(lambda x: np.fromstring(x.strip('[]'), sep=' '))
But for my case, using the embeddings created with the batch, I appended them to the df, as they were created separately, the above did not work. Turn out that the separator should have a `, ` and not ` ` as its value.
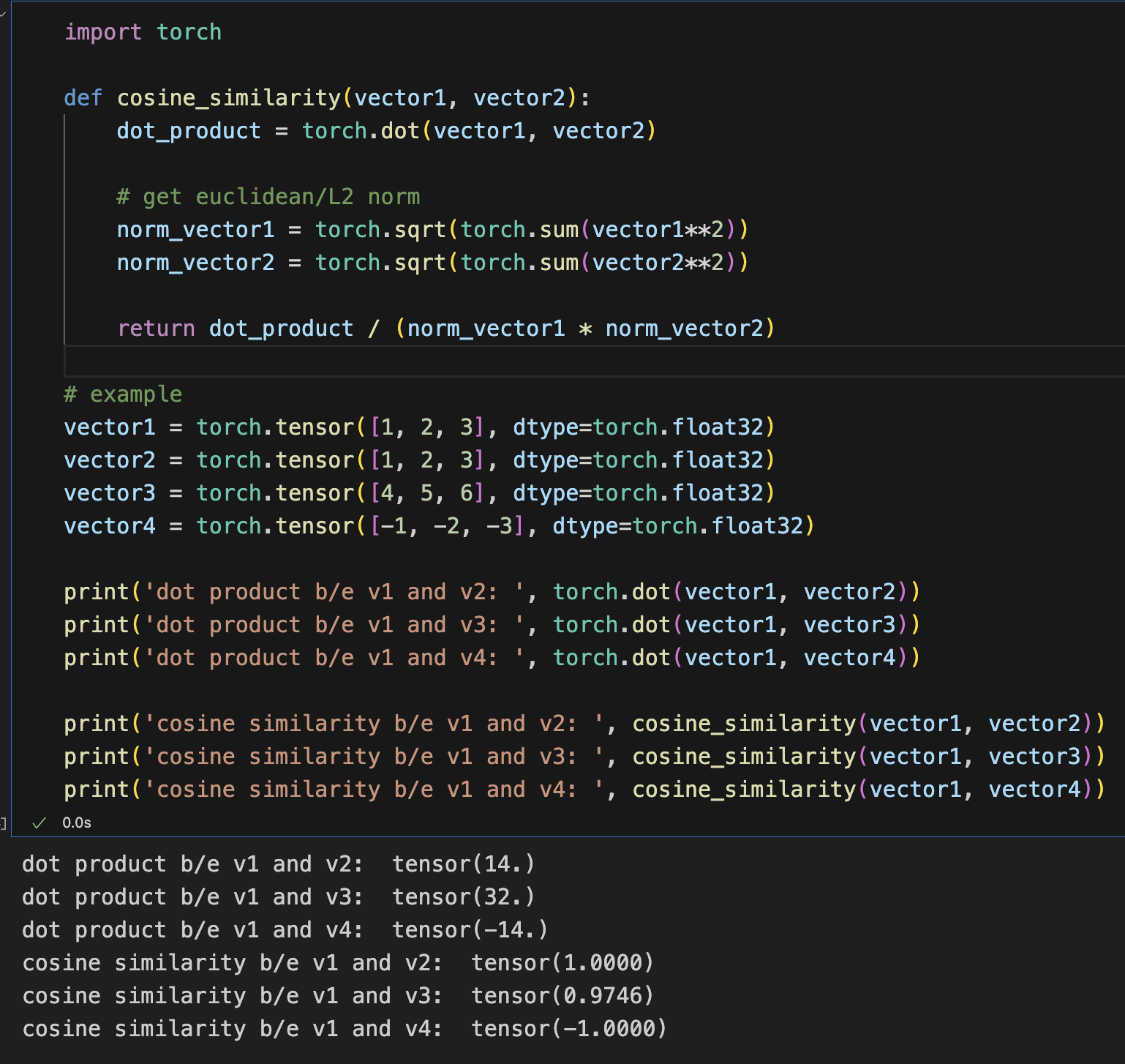
Next, using dot score between a query embedding and the embeddings, we could get a passage from the pdf that was relevant
Next, I learned a bit more about cosine similarity and dot product.The difference is that cosine similarity normalises the dot product over the euclidean distance between the 2 vectors, and if our outputs are already normalised, using just the dot product scores is sufficient (otherwise we do extra computations).Afterwards, came my biggest challenge - getting an LLM to run locally.
First, I loaded `google/gemma-2b-it` and I thought I could run it. I managed to tokenize a sample input, but when I wanted to generate, I got some weird error:
RuntimeError: User specified an unsupported autocast device_type 'mps'
I looked online and one post mentioned that gemma is available for macs, but still I did not understand my issue. Then I decided to freestyle it, and look for an LLM myself on huggingface - I started downloading one of llama's models but it was like 10GB, then I tried one of mistralAI's models and it too was 10GB, the reason I did not download the full models, was because it was getting late, I wanted to see if I can do it with a simple model. So I went to my old friend - gpt-2. All was good, small model, quick download, I was scared that the output might be ... not that good. Unfortunately this fear was correct haha. The output, though talking about nutrition, kept walking around the answer for 256 tokens, but never provided an answer. So I decided to not give up. I started looking more at 'can gemme work on mps?'. And after some videos/blog posts I finally found the answer on this github issue thread. The error that person got was the same as mine, and the torch team responded. Apparently the issue is with the latest version of transformers. If I go back 1 version, or go to a custom dev version the issue is fixes. I went with the latter, and YES! it worked! Or did it ?
Yes, that particular weird runtime error went away, but then a memory issue appeared. My GPU is not strong enough... it said 18GB is max, but more was attempted to be allocated. At that point I accepted my defeat. But nevertheless, I did not want to look for another model now, so I just continued on CPU.
With the prompt:
What are the macronutrients and what are their functions in the body?
It took 1.2 mins to generate 256 token output:
At this point, relief and exhaust took over and I decided to write this blog before I fall asleep on my chair.
Tomorrow I continue, and finish the course.
In addition, another good thing about the teacher is that they provide tips on future upgrades for us to attempt on our own which is awesome.
I forked the github repo and added my file -> here
That is all for today!
See you tomorrow :)